It was an eventful couple of days in Heidelberg at The Identity and Evolution of Cell Types conference: 4 days, around 150 participants, 20 speakers, and a variety of model organisms and research approaches (but mostly single cell RNA sequencing).
Endless forms most beautiful and not so unique
Jumping from mouse brain to cuttlefish cartilage to lamprey neural crest, and even stepping outside of the animal kingdom (and my comfort zone) for a moment to check out some organoids (Gray Camp, Barbara Treutlein), the research painted a picture of the scope of animal diversity and its ancient and complex origins.

Jake Musser’s talk about cell types in sponges (photo credit @tim_wollesen)
A session chaired by Nicole King, starting with her group’s research on choanoflagellates, explored the topic of the origins of multicellularity. In others, moving on to simple multicellular animals, we heard a sponge cell type talk from Jacob Musser (EvoCell’s resident scRNAseq expert), and some views about ctenophore phylogeny from Leonid Moroz (sister to sponges or not? leave your hot take in the comments).
Overall, it seems like we’re all (human, fly, sponge, maybe not ctenophore) relatively similar on the transcriptomic level, and it takes innovative scientific efforts to figure out why and how we then look and work so differently.
The cell tree of life
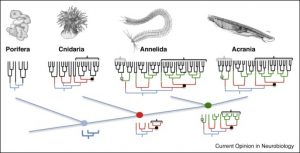
Animals evolved through cell type diversification
(from Arendt et al 2019)
Detlev Arendt’s talk about ”The cell tree of life” introduced the conceptual framework of cell types as ”a set of cells accessing the same regulatory program driving differentiation”. It was interesting to apply that idea to the rest of the talks: much of the research employs single cell technology, which is proving to be a powerful and unbiased way of deciphering the regulatory networks involved in cell type specification.
Sharing is caring is comparing

Closing remarks and discussion panel with chairs and organisers: Stein Aerts, Detlev Arendt, Oliver Hobert, Gunter Wagner and Henrik Kaessman. Lacking representation for women and unicellular organisms in organiser Nicole King’s absence.
The landscape of evolutionary research is changing as we work out new experimental (a variety of scRNAseq, combined with ATAC-seq, methylomes, epitranscriptomes, and functional experiments) and computational approaches. Combining and comparing the many huge datasets from different organisms remains a challenge, and meetings like The Identity and Evolution of Cell Types (first of hopefully many biennial events) are a great platform to establish new collaborations that will drive open science and innovation in evolutionary research.
p.s. We were also there

EvoCell fellows slightly overwhelmed at the computational session. Periklis is unfazed.
Some EvoCell representation at the conference: Periklis talked about neuronal diversity in the sea urchin, Alba had a poster about the cell types in the regenerating crustacean limb, and a couple more of us joined the meeting for bad coffee and a learning experience.

End of Periklis Pagonos’ talk, we’re in the acknowledgements

EvoCell’s Alba Almazan next to her poster

Recent Comments